
Paix, B.; Othmani, A.; Debroas, D.; Culioli, G.; Briand, J. Temporal Covariation of Epibacterial Community and Surface Metabolome in the Mediterranean Seaweed Holobiont Taonia atomaria . Environ Microbiol 2019, 1462-2920.14617. https://doi.org/10.1111/1462-2920.14617.
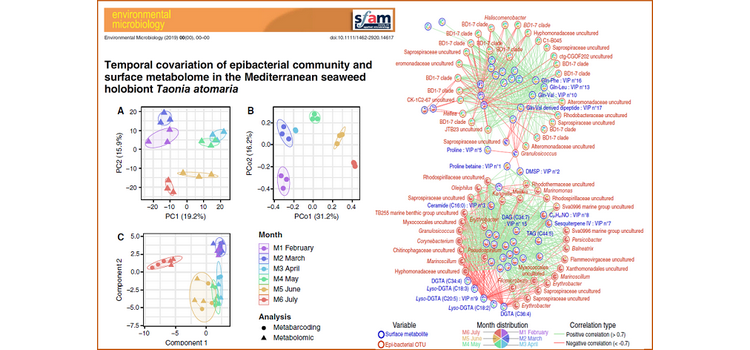
An integrative multi-omics approach allowed monthly variations for a year of the surface metabolome and the epibacterial community of the Mediterranean Phaeophyceae Taonia atomaria to be investigated. The LC–MS-based metabolomics and 16S rDNA metabarcoding data sets were integrated in a multivariate meta-omics analysis (multi-block PLS-DA from the MixOmic DIABLO analysis) showing a strong seasonal covariation (Mantel test: p < 0.01). A network based on positive and negative correlations between the two data sets revealed two clusters of variables, one relative to the ‘spring period’ and a second to the ‘summer period’. The ‘spring period’ cluster was mainly characterized by dipeptides positively correlated with a single bacterial taxon of the Alteromonadaceae family (BD1-7 clade). Moreover, ‘summer’ dominant epibacterial taxa from the second cluster (including Erythrobacteraceae, Rhodospirillaceae, Oceanospirillaceae and Flammeovirgaceae) showed positive correlations with few metabolites known as macroalgal antifouling defences [e.g. dimethylsulphoniopropionate (DMSP) and proline] which exhibited a key role within the correlation network. Despite a core community that represents a significant part of the total epibacteria, changes in the microbiota structure associated with surface metabolome variations suggested that both environment and algal host shape the bacterial surface microbiota.
