Biofilm formation and c-di-GMP signaling in Shewanella oneidensis
Supervisor: MOLMERET Maëlle
Co-supervisor: BARAQUET Claudine
Abstract
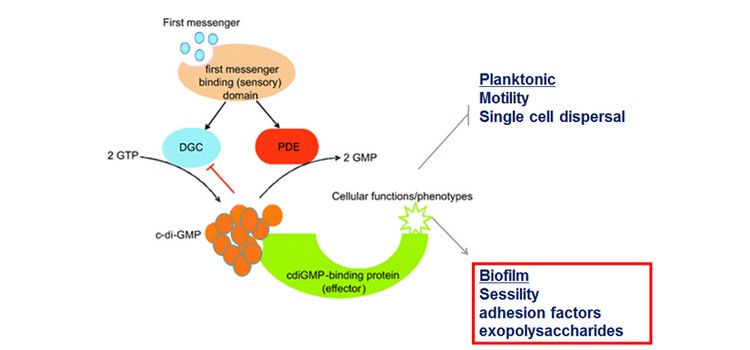
Biofilms are surface-attached bacteria embedded in a self-produced matrix of polysaccharides, proteins and nucleic acids. It can cause economic and environmental drawbacks for marine industries such as ships drag reduction. In many bacteria, biofilm formation is regulated by cyclic-di-guanosine monophosphate (c-di-GMP). It is a ubiquitous second messenger, which affects the transition between planktonic and biofilm lifestyles. High intracellular c-di-GMP is usually associated with biofilm production.
Understanding how marine bacteria adhere and form biofilms on surfaces is crucial to develop new coatings with targeted anti-biofilm properties. We seek to identify, in the model bacteria Shewanella oneidensis, genes involved in biofilm formation using high-throughput transposon insertion sequencing (Tn-seq) and the role of c-di-GMP using RNA-seq.
We constructed a Tn-insertion library in S. oneidensis. We tested different mating conditions to build the library. We also constructed S. oneidensis strains with low or high concentrations of c-di-GMP as well as a strain deleted of flrA. FlrA is homologous to FleQ, a key c-di-GMP responsive transcriptional regulator of biofilm formation in Pseudomonas aeruginosa.
We obtained a transposon mutant library to study biofilm formation in S. oneidensis. The flrA mutant is affected in motility and biofilm formation. As shown by others, c-di-GMP affects biofilm formation in S. oneidensis. We expect, using RNA-seq and Tn-seq experiments, to find new candidates for biofilm inhibition. Because of the widespread conservation of c-di-GMP signaling in bacteria and the absence of encoded associated enzymes in eukaryotic organisms, inhibition of this pathway offers an attractive approach to interfere with biofilm formation.
Keywords
Biofilm, c-di-GMP, high-throughput sequencing, Shewanella oneidensis